Research
Our VALUES
Research benefits from a wide variety of ideas. Therefore,
actively supporting diversity promotes excellence. In our lab, we value
the diversity of backgrounds present in our group, and strive to foster an
environment that promotes equity, diversity, and inclusion from
underrepresented groups in STEM.
Protein synthesis involves the translation of ribonucleic acid information into proteins, the building blocks of life. The initial step of protein synthesis consists of the eukaryotic translation initiation factor 4E (eIF4E) binding to the 5’ cap of mRNAs. However, many cellular stresses repress cap-dependent translation to conserve energy by sequestering eIF4E. This raises a fundamental question in biology as to how proteins are synthesized during periods of cellular stress and eIF4E inhibition. Research in our laboratory will build upon the discovery that protein synthesis is dynamic and a key point of regulation for gene expression. Our lab focuses on a switch in translation initiation to the cap-binding protein eIF4E2 and to the composition of ribosomes to produce a specialized, adaptive protein output in human cells. One of our main focuses will be on cancer because as human tumors display considerable diversity in their genetic makeup, they share common physiological attributes such as a hypoxic microenvironment that contribute to the malignant phenotype. As oxygen only diffuses through a few layers of cells, the vast majority of cancer cells that populate a tumor are thought to be exposed to hypoxic conditions. Therefore, understanding how eIF4E2-dependent translation, a mechanism scarcely used by normal oxygenated cells, functions and contributes to the expression of the tumor cell phenotype will provide unique opportunities for cancer therapy. The long-term goal of our research program is to highlight the plasticity of the translation apparatus that allows human cells to respond to changes in oxygen availability.
Our Projects
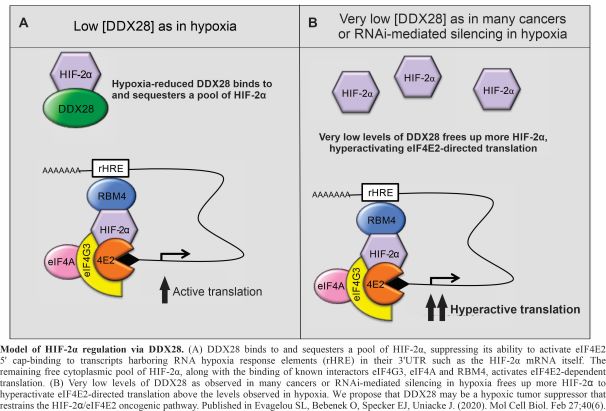
The regulation of HIF2α via the RNA helicase DDX28
In 2019, the Nobel Prize in Physiology or Medicine was awarded to Drs. Kaelin, Ratcliffe, and Semenza for their discoveries of how cells sense and adapt to oxygen availability. These discoveries, first made about three decades prior, center around the essential transcription factors, the Hypoxia Inducible Factors (HIFs). Hypoxia stabilizes HIFα subunits (HIF1α or HIF2α) that each assemble with constitutively expressed HIFα to produce active HIF1 and HIF2 transcription factors. These activate the transcription of genes involved in embryonic development, blood vessel formation, cellular metabolism, wound healing, muscle exercise, and cancer progression. We have identified a new mode of HIF2α regulation whereby the RNA helicase DDX28 binds to and sequesters HIF2α, restraining its ability to activate eiF4E2-directed translation. We continue to explore the mechanisms employed by DDX28 in HIF2α regulation and in the general response to hypoxia using techniques such as mutagenesis, cloning, protein-protein interactions, fluorescence microscopy, and qRT-PCR. DDX28 has been observed to be reduced in patients with advanced cancer (and protective in high amounts) suggesting that it has a role as a tumour suppressor. This project will give us insights into fundamental aspects of cellular adaptation to stress through translation regulation and cancer biology.
The regulation of HIFα and hypoxic translation by ISGylation
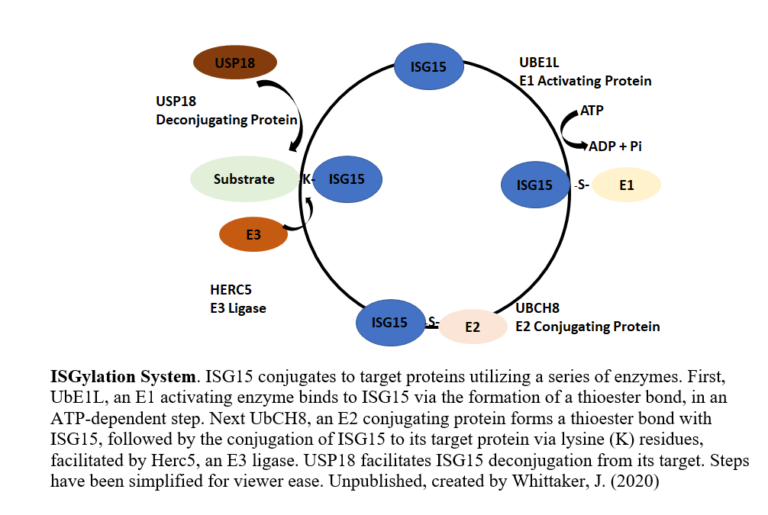
ISG15 is a ubiquitin-like modifier protein that gets covalently bound to a variety of different proteins (with a variety of different effects) or excreted by the cell in response to interferon signaling induced by viruses. Both HIFα and eiF4E2 can be ISGylated, but the effects on hypoxic gene expression are unknown. In this project, we will investigate how ISG15 modifies the activity and cellular localization of HIFα and eIF4E2 through western blot, immunoprecipitation, fluorescence microscopy, polysome fractionation, and qRT-PCR. We will also investigate how hypoxia influences the production of ISG15 and the ISGylation enzymes, as this process may not be solely an antiviral response. Many cancers express high amounts of ISG15 suggesting that it is an oncogene. This project will provide new insights into how hypoxic gene expression can be shaped by the post-translational modification ISG15 cells. This will have applications to fundamental understanding of basic molecular biology and to gene expression strategies hijacked by cancer cells.
The influence of hypoxia on the composition and function of ribosomes
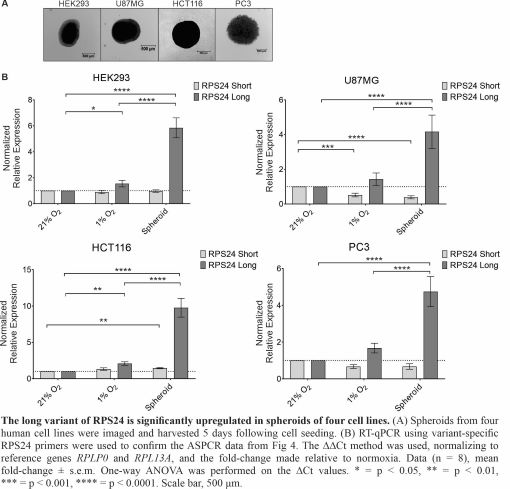
The perception of the ribosome, the nexus of protein synthesis within the cell, has changed dramatically throughout the past couple of decades. No longer viewed as a completely static and passive structure, mounting evidence suggests that cells maintain and/or induce a heterogeneous pool of ribosomes. This nascent field suggests that “specialized ribosomes” have altered transcript specificity, a feature which could be exploited by cancer cells. We provided the first evidence that the composition of human ribosomes and the alternative splicing of ribosomal protein genes changes in response to a stimulus (hypoxia). Specifically, through Tandem Mass Tags mass spectrometry, we identified that ribosomes were twice more likely to contain RPS12 in hypoxia (1% O2) than in normoxia (21% O2) and that these ribosomes initiated the translation of transcripts with long, structured UTRs. We also uncovered a hypoxia-induced alternate splicing event in the RPS24 mRNA that produces distinct long (RPS24L) and short (RPS24S) protein isoforms that can incorporate into ribosomes. Hypoxia increased the RPS24L/RPS24S protein ratio by two-fold in cell monolayers, but over five-fold in 3D cell culture models (spheroids). We will continue to investigate which transcripts are bound to RPS12- and RPS24 (long or short)-containing ribosomes and how this influences cellular adaptation to hypoxia. We will also investigate how RPS24 alternative splicing is regulated in hypoxia and how the RPS24L isoform contributes to hypoxic cell survival. Trainees will learn techniques such as advanced mass spectrometry and bioinformatics analysis, siRNA silencing, hypoxic human cell culture, and qRT-PCR. We noted an increase in the RPS24L/RPS24S isoform ratio in advanced prostate cancer. Therefore, not only will this project contribute to our understanding of ribosomes, an essential molecular machine within the cell, but to a process that may be important to cancer progression.
Published in Brumwell A, Fell L, Obress L, Uniacke J. (2020). RNA. Mar;26(3):361-371.
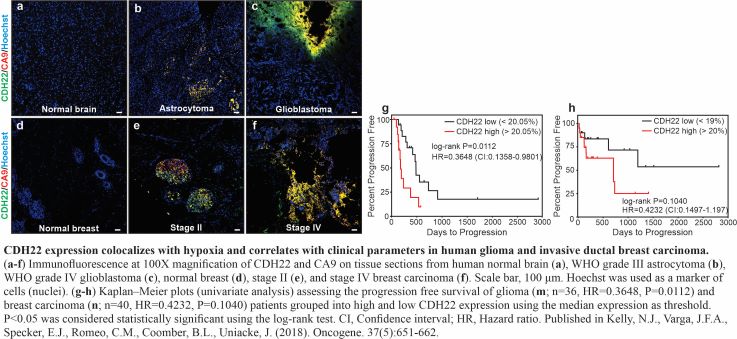
The role of Neuropeptide Y receptors and Cadherin-22 in hypoxic breast cancer cell survival, proliferation, and signaling
In Canada, 10% of new breast cancers are metastatic at diagnosis, and 30% of women diagnosed at early stages develop metastases1. Three general processes contribute to cancer initiation and progression: somatic mutations, epigenetic alterations, and the tumor microenvironment. Somatic mutations and epigenetic alterations draw substantial attention because they are easily assayed by high-throughput molecular techniques. In contrast, changes in the tumor microenvironment are less easily interrogated but have profound effects on gene expression. Hypoxia arises in tumors through oncogene-driven proliferation of cancer cells in the absence of efficient vasculature and is strongly connected to the acquisition of metastatic properties leading to patient mortality2. We identified Neuropeptide peptide Y receptors and cadherin-22 (CDH22), hypoxic cell surface proteins that play roles in signaling and cell migration/invasion of hypoxic breast cancer cells. We propose to investigate the molecular signaling mechanisms and in vivo roles of these receptors in tumors to develop its potential as a therapeutic target against breast cancer metastasis. Students will be trained in imaging techniques in 3D cell culture models and tumors, in vitro cell migration and invasion assays, and molecular techniques such as chromatin immunoprecipitation and qRT-PCR.
Investigating the influence of physiological oxygen on gene expression
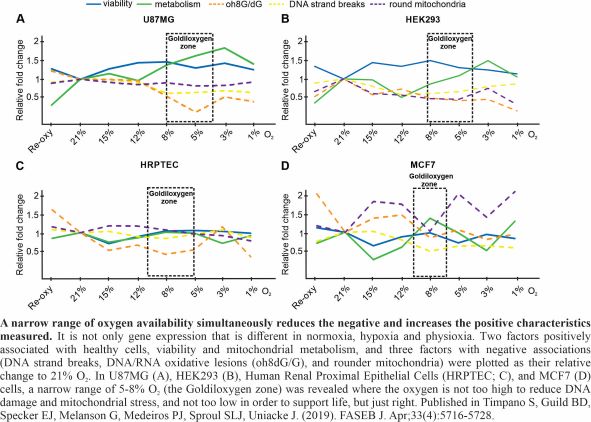
Hypoxia is traditionally defined as 1% O2 availability or less, while normoxia as 21% O2 (ambient air). In human cell culture, oxygen is a surprisingly neglected variable, especially considering that physiological oxygen levels in the human body fall within the range of 1-11% O2 (physioxia) depending on the tissue. Most tissues are in the lower part of that range and the dissolved oxygen within cells is estimated to equate to 1-2% O2. We showed in five different human cell lines that two distinct modes of cap-dependent translation initiation (eIF4E- and eIF4E2-directed) are active during physioxia and act on separate pools of mRNA. This suggests that physioxic cells could have different proteomes than in routine cell culture conditions such as normoxia. In this project, we investigate the mechanisms employed by normoxic, hypoxic and physioxic cells to express their genes at the level of translation. Students will be trained in advanced RNA sequencing techniques, nucleic acid purification and bioinformatics software. This project has the potential to impact cell culture practices and our fundamental knowledge of the role of oxygen in gene expression.